Using Quadtrees
Derek Friend
Last compiled on August 29, 2023
Source:vignettes/quadtree-usage.Rmd
quadtree-usage.RmdVignette content
This vignette covers ways of interacting with a Quadtree
object after it has been created - for example, extracting values and
modifying values.
Retrieving basic info about a quadtree
There are a number of functions for retrieving basic info about a
Quadtree.
Getting a summary of a quadtree
A basic summary of a quadtree can be shown by just typing the
variable name or using the summary() function:
qt
#> class : Quadtree
#> # of cells : 4189
#> min cell size : 250
#> extent : 0, 64000, 0, 64000 (xmin, xmax, ymin, ymax)
#> crs : PROJCRS["unknown",
#> BASEGEOGCRS["unknown",
#> DATUM["Unknown based on Airy 1830 ellipsoid",
#> ELLIPSOID["Airy 1830",6377563.396,299.3249646,
#> LENGTHUNIT["metre",1,
#> ID["EPSG",9001]]]],
#> PRIMEM["Greenwich",0,
#> ANGLEUNIT["degree",0.0174532925199433,
#> ID["EPSG",9122]]]],
#> CONVERSION["Transverse Mercator",
#> METHOD["Transverse Mercator",
#> ID["EPSG",9807]],
#> PARAMETER["Latitude of natural origin",49,
#> ANGLEUNIT["degree",0.0174532925199433],
#> ID["EPSG",8801]],
#> PARAMETER["Longitude of natural origin",-2,
#> ANGLEUNIT["degree",0.0174532925199433],
#> ID["EPSG",8802]],
#> PARAMETER["Scale factor at natural origin",0.9996012717,
#> SCALEUNIT["unity",1],
#> ID["EPSG",8805]],
#> PARAMETER["False easting",400000,
#> LENGTHUNIT["metre",1],
#> ID["EPSG",8806]],
#> PARAMETER["False northing",-100000,
#> LENGTHUNIT["metre",1],
#> ID["EPSG",8807]]],
#> CS[Cartesian,2],
#> AXIS["easting",east,
#> ORDER[1],
#> LENGTHUNIT["metre",1,
#> ID["EPSG",9001]]],
#> AXIS["northing",north,
#> ORDER[2],
#> LENGTHUNIT["metre",1,
#> ID["EPSG",9001]]]]
#> values : 0.109, 0.974 (min, max)Getting the projection of a quadtree
projection() can be used to get and set the projection
of the Quadtree. In this case there is no projection, so an
empty string is returned.
projection(qt)
#> [1] "PROJCRS[\"unknown\",\n BASEGEOGCRS[\"unknown\",\n DATUM[\"Unknown based on Airy 1830 ellipsoid\",\n ELLIPSOID[\"Airy 1830\",6377563.396,299.3249646,\n LENGTHUNIT[\"metre\",1,\n ID[\"EPSG\",9001]]]],\n PRIMEM[\"Greenwich\",0,\n ANGLEUNIT[\"degree\",0.0174532925199433,\n ID[\"EPSG\",9122]]]],\n CONVERSION[\"Transverse Mercator\",\n METHOD[\"Transverse Mercator\",\n ID[\"EPSG\",9807]],\n PARAMETER[\"Latitude of natural origin\",49,\n ANGLEUNIT[\"degree\",0.0174532925199433],\n ID[\"EPSG\",8801]],\n PARAMETER[\"Longitude of natural origin\",-2,\n ANGLEUNIT[\"degree\",0.0174532925199433],\n ID[\"EPSG\",8802]],\n PARAMETER[\"Scale factor at natural origin\",0.9996012717,\n SCALEUNIT[\"unity\",1],\n ID[\"EPSG\",8805]],\n PARAMETER[\"False easting\",400000,\n LENGTHUNIT[\"metre\",1],\n ID[\"EPSG\",8806]],\n PARAMETER[\"False northing\",-100000,\n LENGTHUNIT[\"metre\",1],\n ID[\"EPSG\",8807]]],\n CS[Cartesian,2],\n AXIS[\"easting\",east,\n ORDER[1],\n LENGTHUNIT[\"metre\",1,\n ID[\"EPSG\",9001]]],\n AXIS[\"northing\",north,\n ORDER[2],\n LENGTHUNIT[\"metre\",1,\n ID[\"EPSG\",9001]]]]"Getting the number of cells in a quadtree
n_cells() returns the number of cells in the quadtree.
It has one optional parameter, terminal_only - if
TRUE, the number of cells of the quadtree that are terminal
(i.e. have no children) is returned. If FALSE, the total
number of nodes in the quadtree is returned.
Getting the extent of a quadtree
extent() can be used to return the extent of the
quadtree. It has one optional parameter original - if
FALSE (the default), it returns the total extent covered by
the quadtree. If TRUE, the function returns the extent of
the original raster used to create the quadtree, before NA
rows/columns were added to pad the dimensions. You may need to preface
extent() with quadtree:: to avoid conflicts
with the raster package.
Retrieving cell-level data
Extracting values
Values can be ‘extracted’ at point locations using the
extract() function. This function has one optional
parameter - extents. If extents is
FALSE (the default), then only the values at the point
locations are returned. If extents is TRUE,
then a matrix is returned that also returns the x and y limits of each
cell in addition to the cell value.
pts <- cbind(x = c(5609, 3959, 20161, 27662, 32763),
y = c(10835, 29586, 31836, 10834, 36337))
plot(qt, crop = TRUE, border_lwd = .3, na_col = NULL)
points(pts, pch = 16)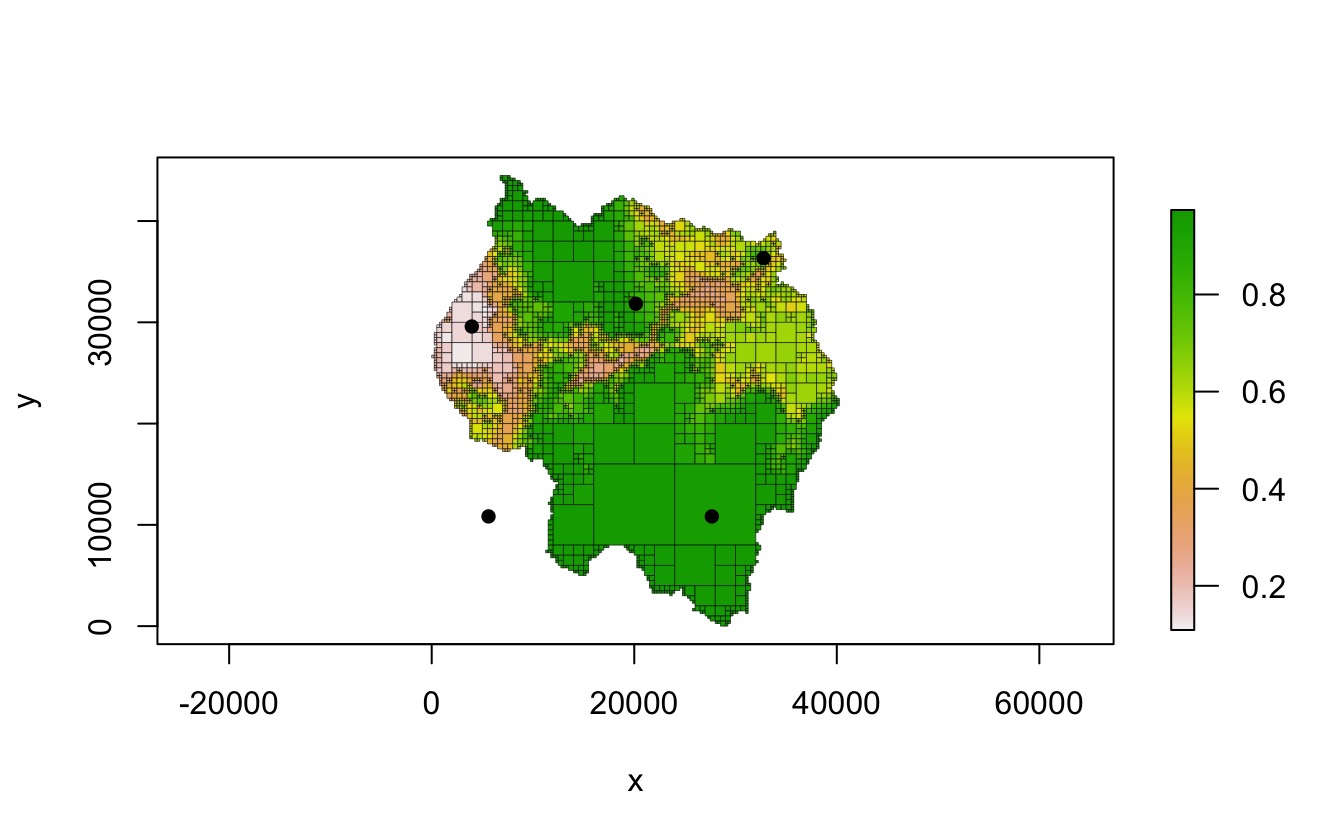
quadtree::extract(qt, pts)
#> [1] NaN 0.1451094 0.8202500 0.9556426 0.6800000
quadtree::extract(qt, pts, extents = TRUE)
#> id xmin xmax ymin ymax value
#> [1,] 4315 0 8000 8000 16000 NaN
#> [2,] 1814 2000 4000 28000 30000 0.1451094
#> [3,] 3378 20000 21000 31000 32000 0.8202500
#> [4,] 4545 24000 32000 8000 16000 0.9556426
#> [5,] 1555 32500 33000 36000 36500 0.6800000Retrieving neighbor relationships
In addition to extracting values from cells, it is possible to
determine neighbor relationships between cells via the
get_neighbors() function. Given a point,
get_neighbors() returns the cells that neighbor the cell
that the point falls in. Note that cells that are diagonal from each
either (i.e. only touch at the corner) are considered to be
neighbors.
get_neighbors(qt, as.numeric(pts[3,]))
#> id xmin xmax ymin ymax value
#> [1,] 967 19500 20000 32000 32500 0.8557500
#> [2,] 1038 20000 21000 32000 33000 0.8236875
#> [3,] 1039 21000 22000 32000 33000 0.7893750
#> [4,] 3279 19000 20000 31000 32000 0.9098125
#> [5,] 3285 19000 20000 30000 31000 0.9466250
#> [6,] 3380 20000 21000 30000 31000 0.8688750
#> [7,] 3379 21000 22000 31000 32000 0.8061875
#> [8,] 3381 21000 22000 30000 31000 0.8125625Copying a quadtree
A key concept to understand is that Quadtree objects are
pointers to C++ objects, and copying these objects simply by
assigning them to a new variable makes a shallow copy rather
than a deep copy. This is important to note because this
differs from how R objects normally behave. For example, if I have a
data frame named df1, I can make a copy of it by doing
df2 <- df1. This makes a deep copy of
df1 - if I were to modify df2,
df1 would remain unchanged. This is not the case with
Quadtree objects. The R object that users interact with is
a pointer to a C++ object. If I were to attempt to make copy of a
quadtree qt1 by doing qt2 <- qt1, this
simply copies the pointer to the C++ object, so
qt1 and qt2 point to the same object. Thus, if
qt2 is modified (for example, by using a function like
set_values() or transform_values()),
qt1 will also be modified.
The copy() function can be used to make a deep copy of a
Quadtree object. This is useful if the user wishes to make
changes to a quadtree without modifying the original.
qt_copy <- copy(qt)
qt
#> class : Quadtree
#> # of cells : 4189
#> min cell size : 250
#> extent : 0, 64000, 0, 64000 (xmin, xmax, ymin, ymax)
#> crs : PROJCRS["unknown",
#> BASEGEOGCRS["unknown",
#> DATUM["Unknown based on Airy 1830 ellipsoid",
#> ELLIPSOID["Airy 1830",6377563.396,299.3249646,
#> LENGTHUNIT["metre",1,
#> ID["EPSG",9001]]]],
#> PRIMEM["Greenwich",0,
#> ANGLEUNIT["degree",0.0174532925199433,
#> ID["EPSG",9122]]]],
#> CONVERSION["Transverse Mercator",
#> METHOD["Transverse Mercator",
#> ID["EPSG",9807]],
#> PARAMETER["Latitude of natural origin",49,
#> ANGLEUNIT["degree",0.0174532925199433],
#> ID["EPSG",8801]],
#> PARAMETER["Longitude of natural origin",-2,
#> ANGLEUNIT["degree",0.0174532925199433],
#> ID["EPSG",8802]],
#> PARAMETER["Scale factor at natural origin",0.9996012717,
#> SCALEUNIT["unity",1],
#> ID["EPSG",8805]],
#> PARAMETER["False easting",400000,
#> LENGTHUNIT["metre",1],
#> ID["EPSG",8806]],
#> PARAMETER["False northing",-100000,
#> LENGTHUNIT["metre",1],
#> ID["EPSG",8807]]],
#> CS[Cartesian,2],
#> AXIS["easting",east,
#> ORDER[1],
#> LENGTHUNIT["metre",1,
#> ID["EPSG",9001]]],
#> AXIS["northing",north,
#> ORDER[2],
#> LENGTHUNIT["metre",1,
#> ID["EPSG",9001]]]]
#> values : 0.109, 0.974 (min, max)
qt_copy
#> class : Quadtree
#> # of cells : 4189
#> min cell size : 250
#> extent : 0, 64000, 0, 64000 (xmin, xmax, ymin, ymax)
#> crs : PROJCRS["unknown",
#> BASEGEOGCRS["unknown",
#> DATUM["Unknown based on Airy 1830 ellipsoid",
#> ELLIPSOID["Airy 1830",6377563.396,299.3249646,
#> LENGTHUNIT["metre",1,
#> ID["EPSG",9001]]]],
#> PRIMEM["Greenwich",0,
#> ANGLEUNIT["degree",0.0174532925199433,
#> ID["EPSG",9122]]]],
#> CONVERSION["Transverse Mercator",
#> METHOD["Transverse Mercator",
#> ID["EPSG",9807]],
#> PARAMETER["Latitude of natural origin",49,
#> ANGLEUNIT["degree",0.0174532925199433],
#> ID["EPSG",8801]],
#> PARAMETER["Longitude of natural origin",-2,
#> ANGLEUNIT["degree",0.0174532925199433],
#> ID["EPSG",8802]],
#> PARAMETER["Scale factor at natural origin",0.9996012717,
#> SCALEUNIT["unity",1],
#> ID["EPSG",8805]],
#> PARAMETER["False easting",400000,
#> LENGTHUNIT["metre",1],
#> ID["EPSG",8806]],
#> PARAMETER["False northing",-100000,
#> LENGTHUNIT["metre",1],
#> ID["EPSG",8807]]],
#> CS[Cartesian,2],
#> AXIS["easting",east,
#> ORDER[1],
#> LENGTHUNIT["metre",1,
#> ID["EPSG",9001]]],
#> AXIS["northing",north,
#> ORDER[2],
#> LENGTHUNIT["metre",1,
#> ID["EPSG",9001]]]]
#> values : 0.109, 0.974 (min, max)In the next section (“Modifying cell values”), this function is used in the examples to create copies of a quadtree before modifying it so that the original remains unchanged.
Modifying cell values
Two functions are provided for modifying cell values:
set_values() and transform_values().
Setting values of cells
set_values() takes a matrix of points and associated
values and changes the values of the cells the points fall in. Note that
only the values of the terminal cells that the point falls in
are changed.
qt2 <- copy(qt)
set_values(qt2, pts, rep(2, nrow(pts)))
plot(qt2, crop = TRUE, border_lwd = .3)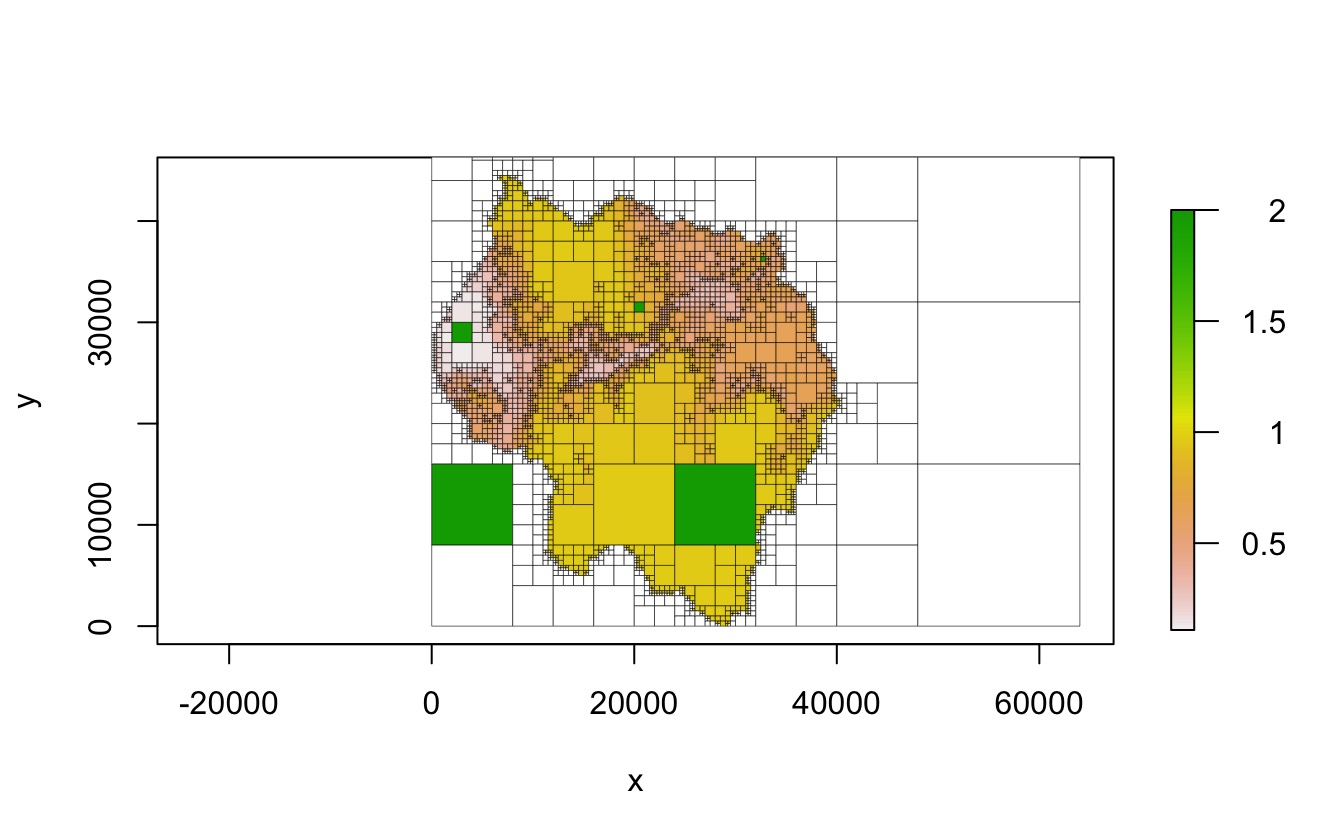
Transforming all cell values
transform_values() uses a function to change all cell
values. For example, we could use this function to cube all of the cell
values:
qt3 <- copy(qt)
transform_values(qt3, function(x) x^3)
par(mfrow = c(1,2))
plot(qt, crop = TRUE, na_col = NULL, border_lwd = .3, zlim = c(0, 1),
legend = FALSE, main = "original quadtree")
plot(qt3, crop = TRUE, na_col = NULL, border_lwd = .3, zlim = c(0, 1),
legend = FALSE, main = "values cubed")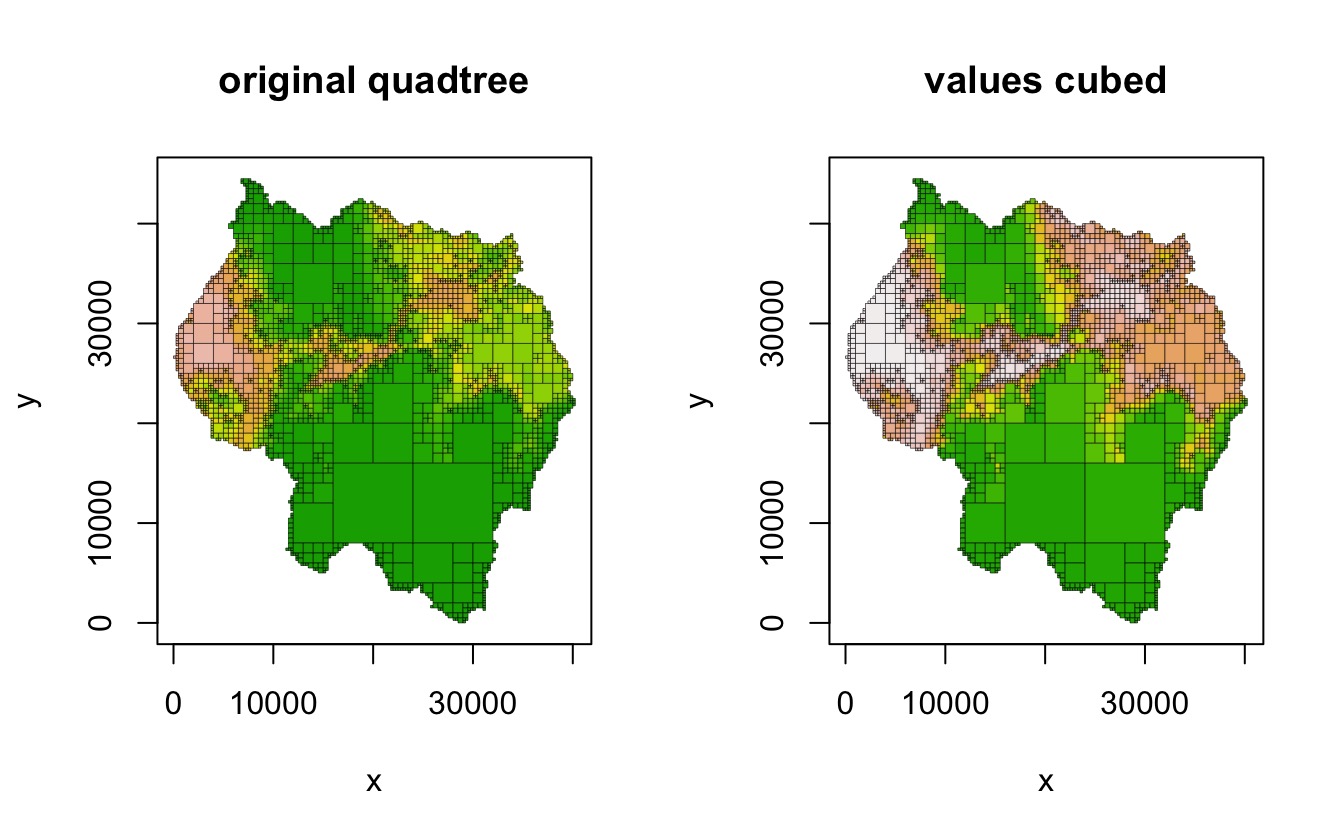
Reading and writing quadtrees
One of the disadvantages of using C++ classes via Rcpp is that the
objects are not preserved between sessions. For example, we could save a
Quadtree to a RData file using
save(), but when we loaded it back in we would get an
error.
qt_temp <- copy(qt)
filepath <- tempfile()
save(qt_temp, file = filepath)
load(filepath)
qt_temp
#> Error in (function (cond) : error in evaluating the argument 'x' in selecting a method for function 'ext': external pointer is not validThe quadtree package provides read and write
functionality via the cereal C++ library, which is used to
serialize C++ objects. read_quadtree() and
write_quadtree() can be used to read and write
Quadtree objects.
filepath <- tempfile()
write_quadtree(filepath, qt)
qt_read <- read_quadtree(filepath)
qt_read
#> class : Quadtree
#> # of cells : 4189
#> min cell size : 250
#> extent : 0, 64000, 0, 64000 (xmin, xmax, ymin, ymax)
#> crs : PROJCRS["unknown",
#> BASEGEOGCRS["unknown",
#> DATUM["Unknown based on Airy 1830 ellipsoid",
#> ELLIPSOID["Airy 1830",6377563.396,299.3249646,
#> LENGTHUNIT["metre",1,
#> ID["EPSG",9001]]]],
#> PRIMEM["Greenwich",0,
#> ANGLEUNIT["degree",0.0174532925199433,
#> ID["EPSG",9122]]]],
#> CONVERSION["Transverse Mercator",
#> METHOD["Transverse Mercator",
#> ID["EPSG",9807]],
#> PARAMETER["Latitude of natural origin",49,
#> ANGLEUNIT["degree",0.0174532925199433],
#> ID["EPSG",8801]],
#> PARAMETER["Longitude of natural origin",-2,
#> ANGLEUNIT["degree",0.0174532925199433],
#> ID["EPSG",8802]],
#> PARAMETER["Scale factor at natural origin",0.9996012717,
#> SCALEUNIT["unity",1],
#> ID["EPSG",8805]],
#> PARAMETER["False easting",400000,
#> LENGTHUNIT["metre",1],
#> ID["EPSG",8806]],
#> PARAMETER["False northing",-100000,
#> LENGTHUNIT["metre",1],
#> ID["EPSG",8807]]],
#> CS[Cartesian,2],
#> AXIS["easting",east,
#> ORDER[1],
#> LENGTHUNIT["metre",1,
#> ID["EPSG",9001]]],
#> AXIS["northing",north,
#> ORDER[2],
#> LENGTHUNIT["metre",1,
#> ID["EPSG",9001]]]]
#> values : 0.109, 0.974 (min, max)Converting quadtrees to other data types
Functions are also provided for converting a Quadtree to
different data types.
Get all cell values as a vector
as_vector() returns the cell values as a numeric vector.
It has one optional parameter - terminal_only. If
TRUE (the default), only the values of the terminal cells
are returned. If FALSE, the values of all cells (including
those with children) are returned.
vec1 <- as_vector(qt)
length(vec1)
#> [1] 4189
summary(vec1)
#> Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
#> 0.1090 0.4843 0.6279 0.6364 0.8142 0.9740 832
vec2 <- as_vector(qt, FALSE)
length(vec2)
#> [1] 5585
summary(vec2)
#> Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
#> 0.1090 0.4910 0.6330 0.6447 0.8320 0.9740 832Convert to a data frame
as_data_frame() converts the quadtree to a data frame.
In the resulting data frame, each row represents a single cell and has
columns describing the cell ID, x and y limits, value, level (i.e. node
depth), the size of its smallest child, the ID of its parent, and
whether it has children. As with as_vector(), there is one
optional parameter called terminal_only - when
TRUE (the default) only the terminal cells are returned,
and when FALSE, all cells are returned.
df1 <- as_data_frame(qt)
dim(df1)
#> [1] 4189 10
head(df1)
#> id hasChildren level xmin xmax ymin ymax value smallestChildLength
#> 3 2 0 2 0 16000 48000 64000 NaN 16000
#> 4 3 0 2 16000 32000 48000 64000 NaN 16000
#> 7 6 0 4 0 4000 44000 48000 NaN 4000
#> 9 8 0 5 4000 6000 46000 48000 NaN 2000
#> 10 9 0 5 6000 8000 46000 48000 NaN 2000
#> 11 10 0 5 4000 6000 44000 46000 NaN 2000
#> parentID
#> 3 1
#> 4 1
#> 7 5
#> 9 7
#> 10 7
#> 11 7
df2 <- as_data_frame(qt, FALSE)
dim(df2)
#> [1] 5585 10
head(df2)
#> id hasChildren level xmin xmax ymin ymax value smallestChildLength
#> 1 0 1 0 0 64000 0 64000 0.7559473 250
#> 2 1 1 1 0 32000 32000 64000 0.7309282 250
#> 3 2 0 2 0 16000 48000 64000 NaN 16000
#> 4 3 0 2 16000 32000 48000 64000 NaN 16000
#> 5 4 1 2 0 16000 32000 48000 0.8131046 250
#> 6 5 1 3 0 8000 40000 48000 0.9623516 250
#> parentID
#> 1 -1
#> 2 0
#> 3 1
#> 4 1
#> 5 1
#> 6 4Convert to a raster
A Quadtree can also be converted to a raster using the
as_raster() function. This function has one optional
parameter named rast, which is used as the template for the
output raster. If NULL (the default), a raster is
automatically created, where the quadtree extent is used as the raster
extent and the smallest cell in the quadtree is used to determine the
resolution of the raster. Note that the value of a raster cell is
determined by the value of the quadtree cell located at the centroid of
the raster cell - thus, if a raster cell overlaps several quadtree
cells, whichever quadtree cell the centroid of the raster cell falls in
will determine the raster cell’s value. If the default value for
rast is used, the raster cells will never overlap with more
than one quadtree cell.